Please, read “BEFORE RUNNING ANY EXPERIMENT” before running ZGPR.
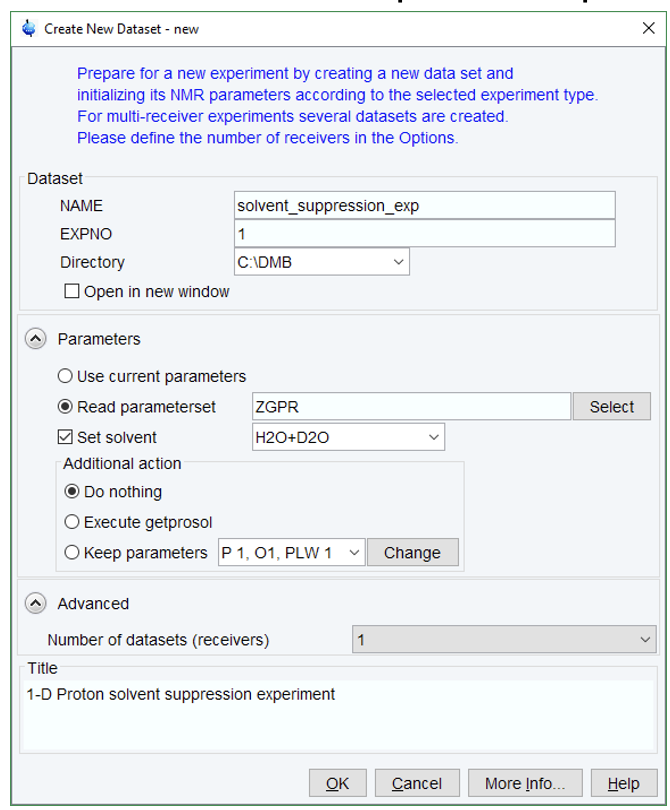
- Create a new dataset with edc or new.
- In the Parameters group
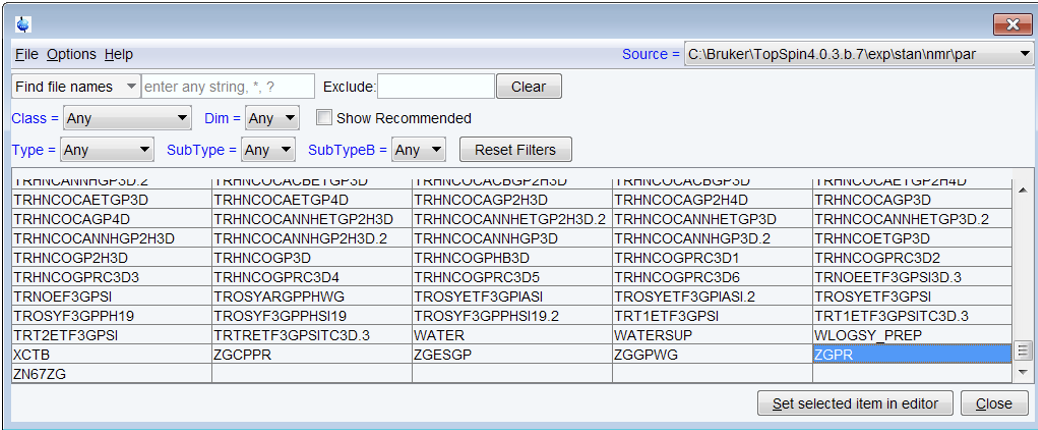
 click Select to open the Experiment Table window (you can just type ZGPR).
click Select to open the Experiment Table window (you can just type ZGPR).
- Select the experiment ZGPR from the table and click on Set selected item in editor.
- Type “getprosol 1H “pulse length” “power in dB” to load the pulses and powers saved on the prosol and also keep your calculated p1.
Ex1: CRP 800 MHz (getprosol 1H 10 -9.38);
Ex2: CRP 700 MHz (getprosol 1H 10 -9.72).
10 is the pulse length in µs – calibrated using pulsecal.
Presaturation experiments are recorded without sample spinning – NEVER spin a sample using a cryoprobe.
- Minimum number of scans ns=8 – if you sample is concentrated, use NS = 8 – 64. If you observe a low signal to noise, increase the NS as necessary.
- TD – Number of points dictate the duration of your FID. For small molecules (32 – 64 k) – have a long T2 relaxation; peptides (16 – 32 k); proteins (2 – 8 k) – short T2 relaxation time.
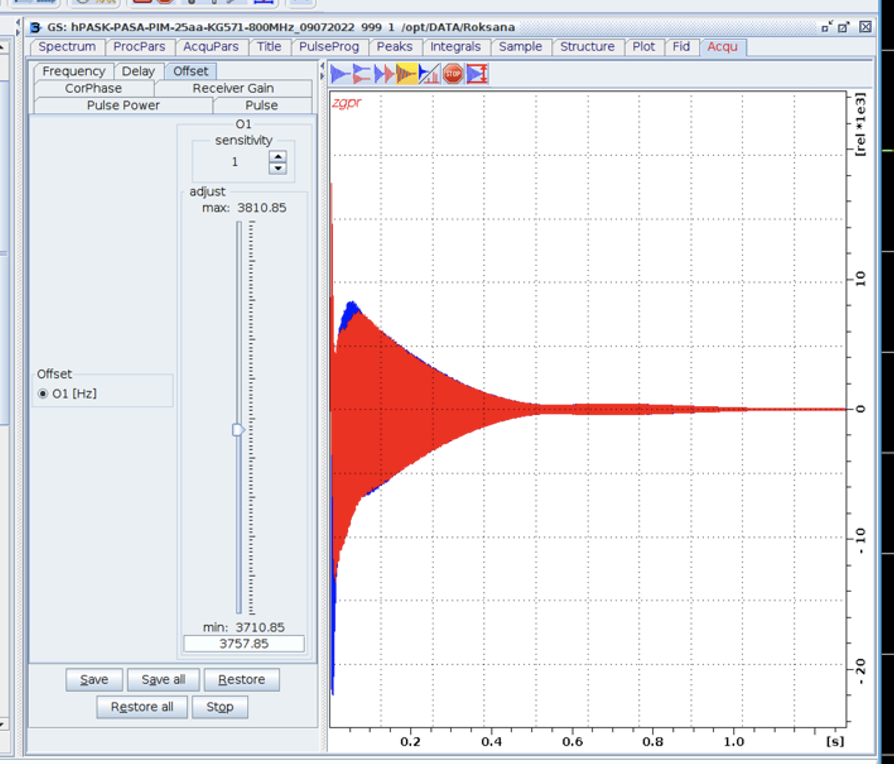
- Set o1p on-resonance to the solvent resonance to be presaturated (o1 may be finely adjusted in gs mode). 4.7 ppm (H2O + D2O).
- Type gs and choose Offset if you want to adjust this value:
- ALWAYS CLICK STOP TO CLOSE gs
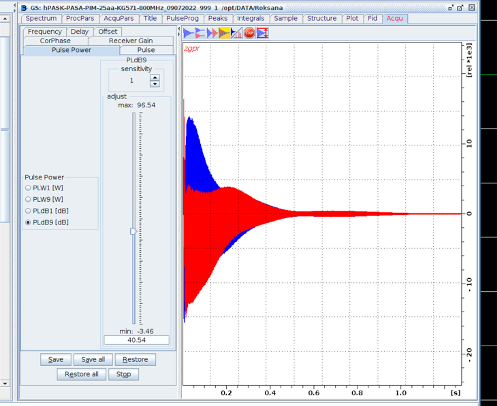
- The power level of the presaturation is defined by PLdB9(start with the power that you get after running pulsecal – usually 45 dB) and the duration d1 (1–4 s), respectively. Adjust PLdB9 using gs (see picture below).
- Calculate the gain by typing rga – after the optimization of the water suppression.
- To start acquisition: zg
- The expected experimental time is displayed with the expt command).
PROCESSING
The recorded data is Fourier transformed with ft (or ef) and phase and baseline corrections are performed using apk and abs, respectively. For proteins, better use qsin combined with ssb=2.
Obs: pre-saturation can also reduce the signal intensities of exchangeable protons. For this reason, other schemes, as the WATERGATE, WET and Excitation Sculpting schemes, can be used to overcome this problem.