Binding observing the Ligand
Theory: soon….
How to set an STD experiment?
1) Adjust everything at zgesgp (water suppression..);
2) wrpa 2 –> re 2;
3) rpar SCREEN_STD all (stddiffesgp.3);
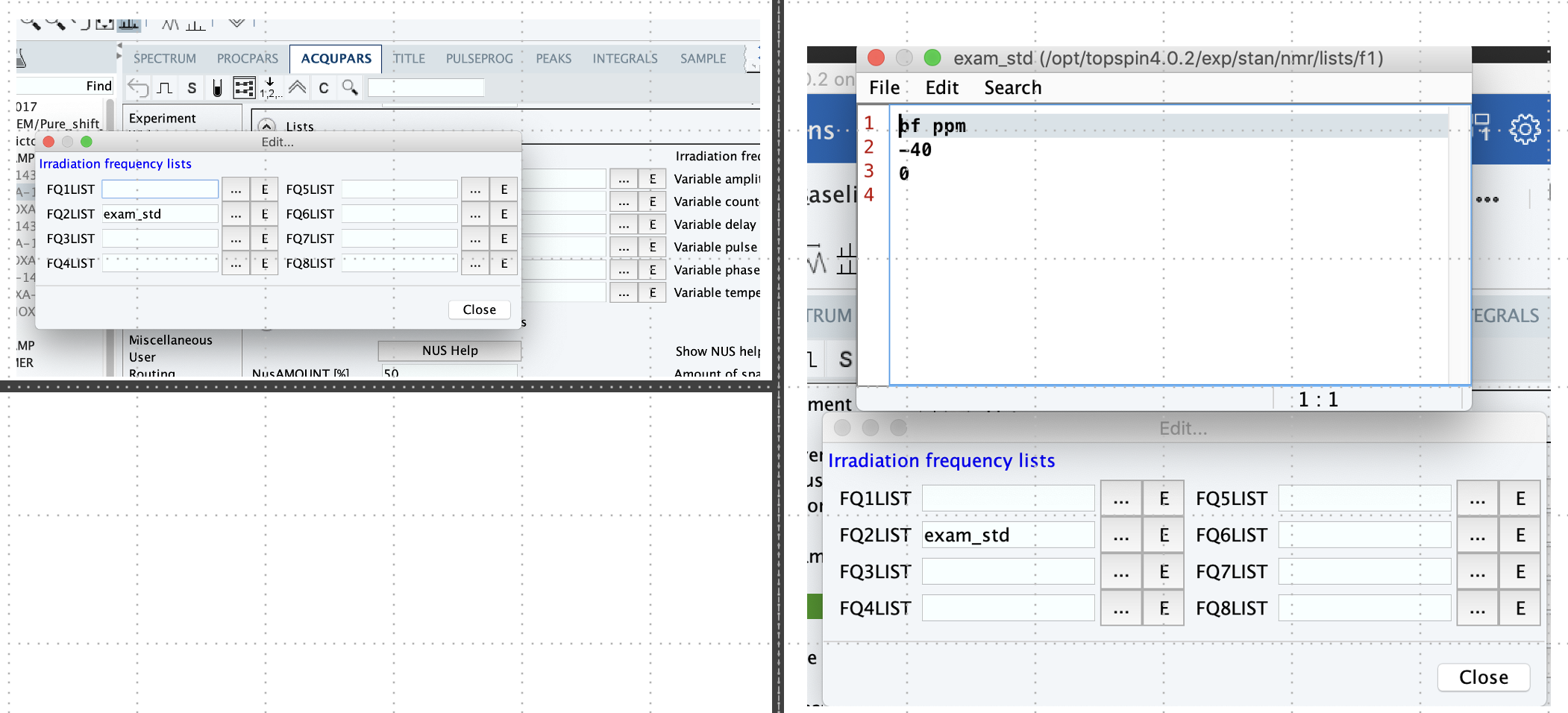
4) Acqupars –> Lists à FQLIST –> edit à FQ2LIST –> E (edit); … (select);
Obs: 1) Reference (ex: -40, 40, 50, or 100 ppm –> avoid chemical shifts close to zero);
2) Protein saturation – Be careful not to saturate any ligand peak.
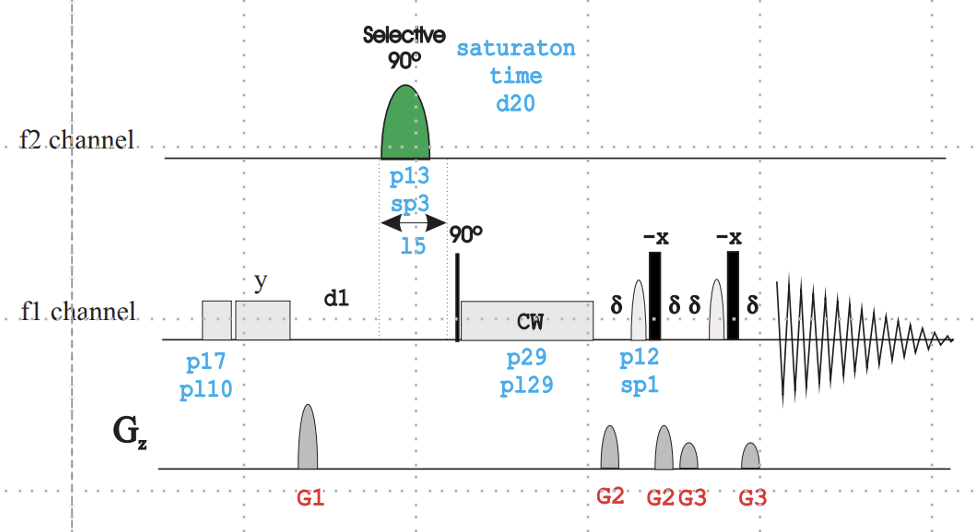
The pulse sequence above contains important parameters that you can play with:
- d20: saturation time [Usually 0.5 – 5 sec] –> 2 sec is a good starting point
Important: The minimum value of d1 is d20, because DELTA1=d1-d31 and d31=d20. I like using d1 = d20.
- d29: spinlock time [10 – 50 msec] – 30 msec is a good starting point
- sp9 : f2 channel – shaped pulse for saturation [40 – 60 dB] or cnst62: 20-200 Hz if ZGOPTNS (-DCALC_POWER)
- NBL: NBL = number of irradiation frequencies (usually NBL = 2)
Processing
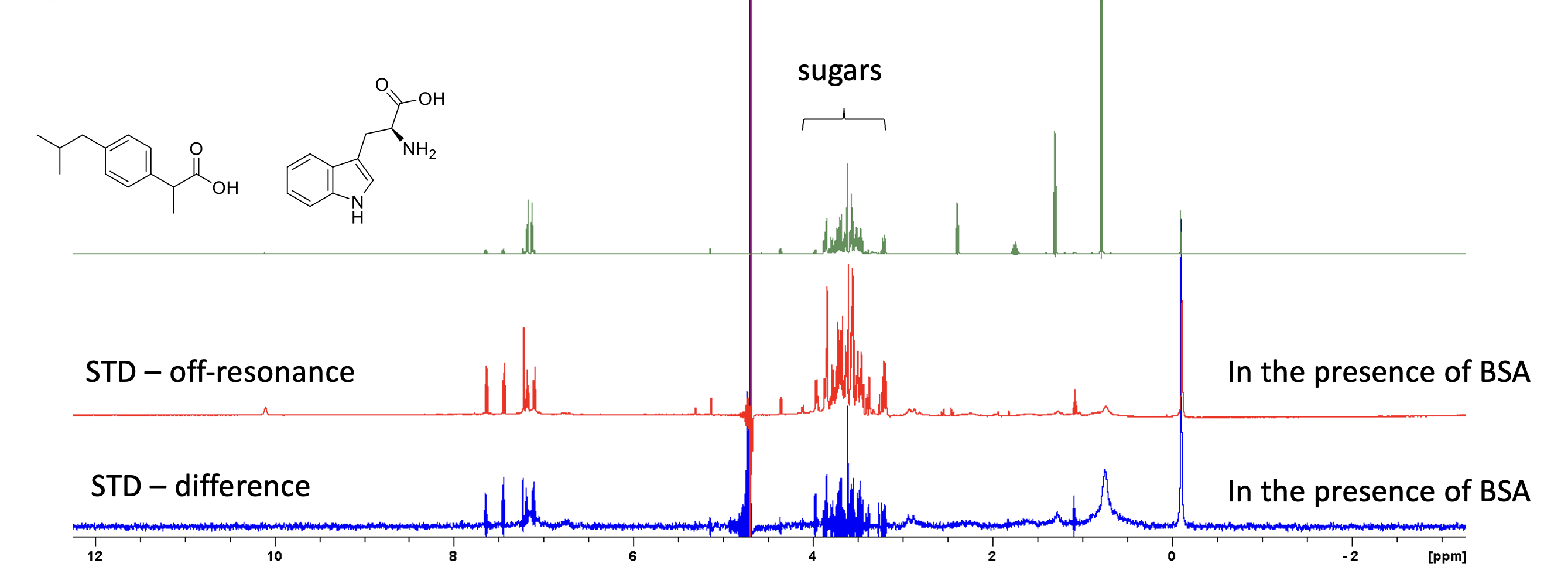
- Type stdsplit – au program for processing (splits your data in 2: reference (STD off-resonance), and difference spectrum (reference – on-resonance spectrum). Process your reference and difference spectrum as a 1D.
–> 20 µM BSA + 400 µM ibuprofen (from a pill) + 400 µM tryptophan. The first experiment is a 1D 1H.
References:
Methods Mol Biol. 2009;534:375-86. doi: 10.1007/978-1-59745-022-5_26.